Sophia Tang

sophtang [at] seas.upenn.edu
Hi! I am a third-year undergraduate student at the University of Pennsylvania conducting research in generative ML for scientific discovery. Aside from research, I study computer science and statistics in the Jerome Fisher Program in Management & Technology.
Currently, I’m part of the Chatterjee Lab, developing theoretical ML frameworks for biological design, and the Mitchell Lab, designing lipid nanoparticle (LNP)-based delivery vehicles to transport mRNA across the blood-brain barrier.
My research ranges from developing theoretical Schrödinger bridge frameworks for generative modelling of branching and interacting particle systems to multi-objective RL and guidance techniques for discrete diffusion - but I’m always exploring new theoretical ideas and thinking about interesting problems to apply them to!
I also write technical articles on my Substack, Alchemy Bio. My most read article, “A Complete Guide to Spherical Equivariant Graph Transformers” (now released on arXiv) is a self-contained guide (99 pages, 46 figures) breaking down the theory behind spherical equivariant graph neural networks (EGNNs) for geometric deep learning.
If you’re also passionate about any of these topics, I would love to connect via LinkedIn or X.
Updates
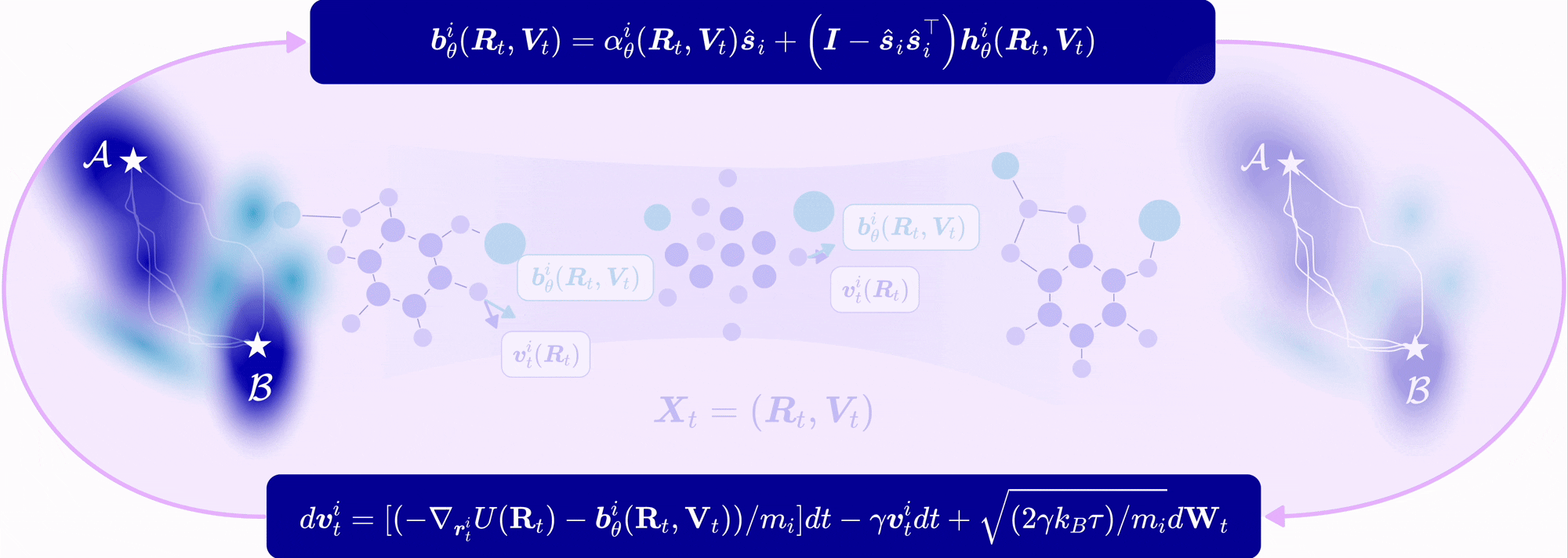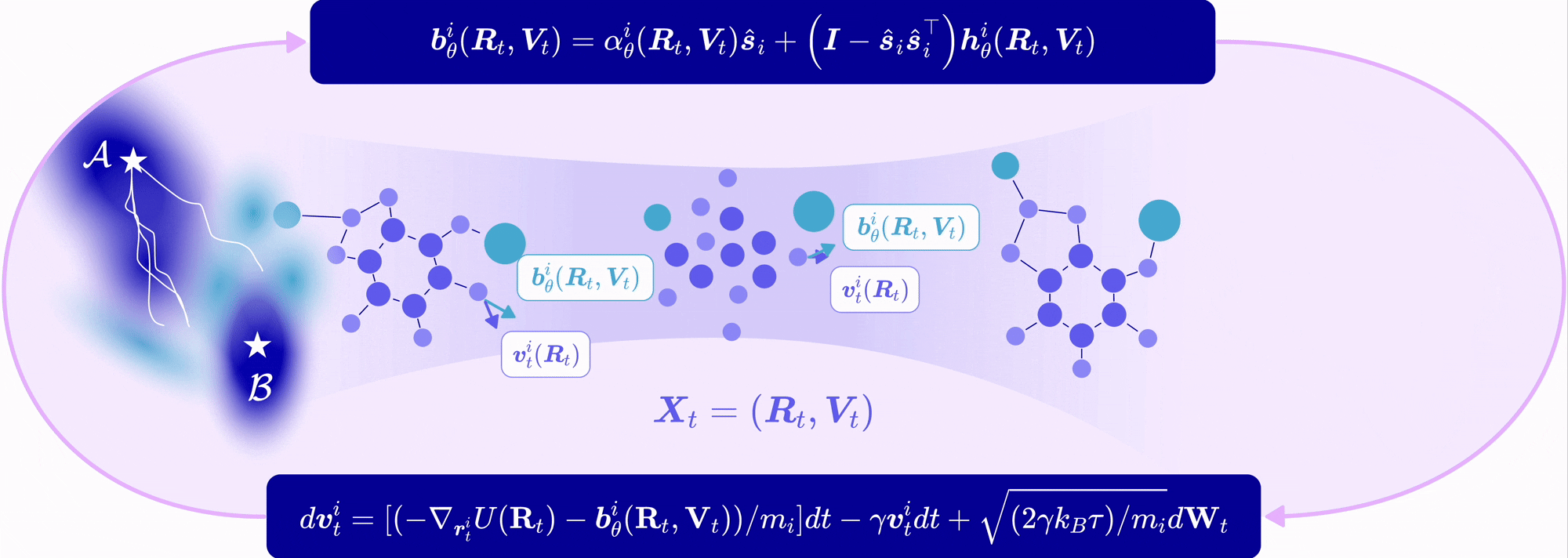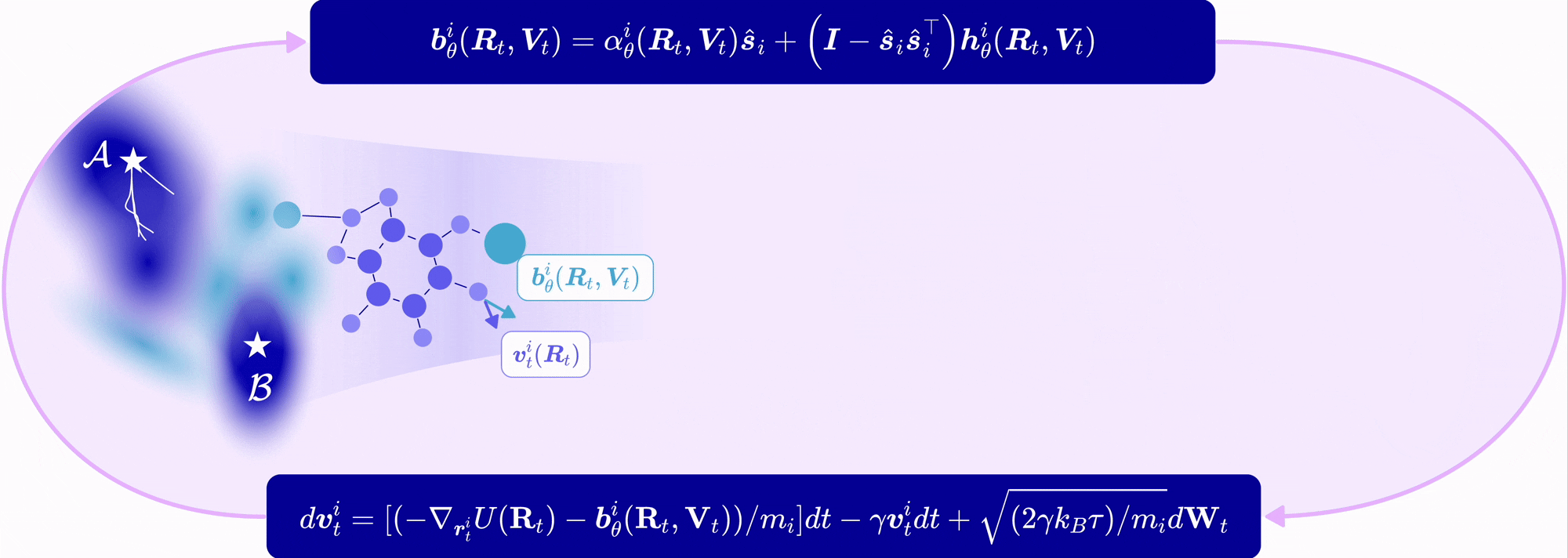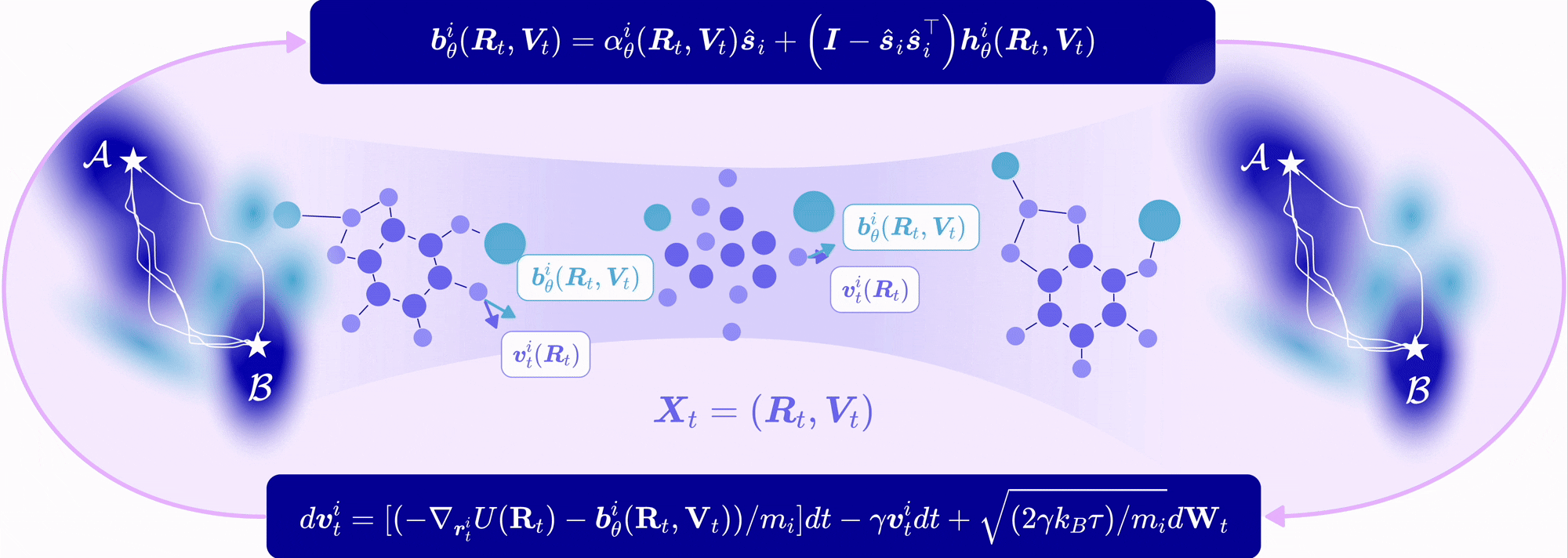
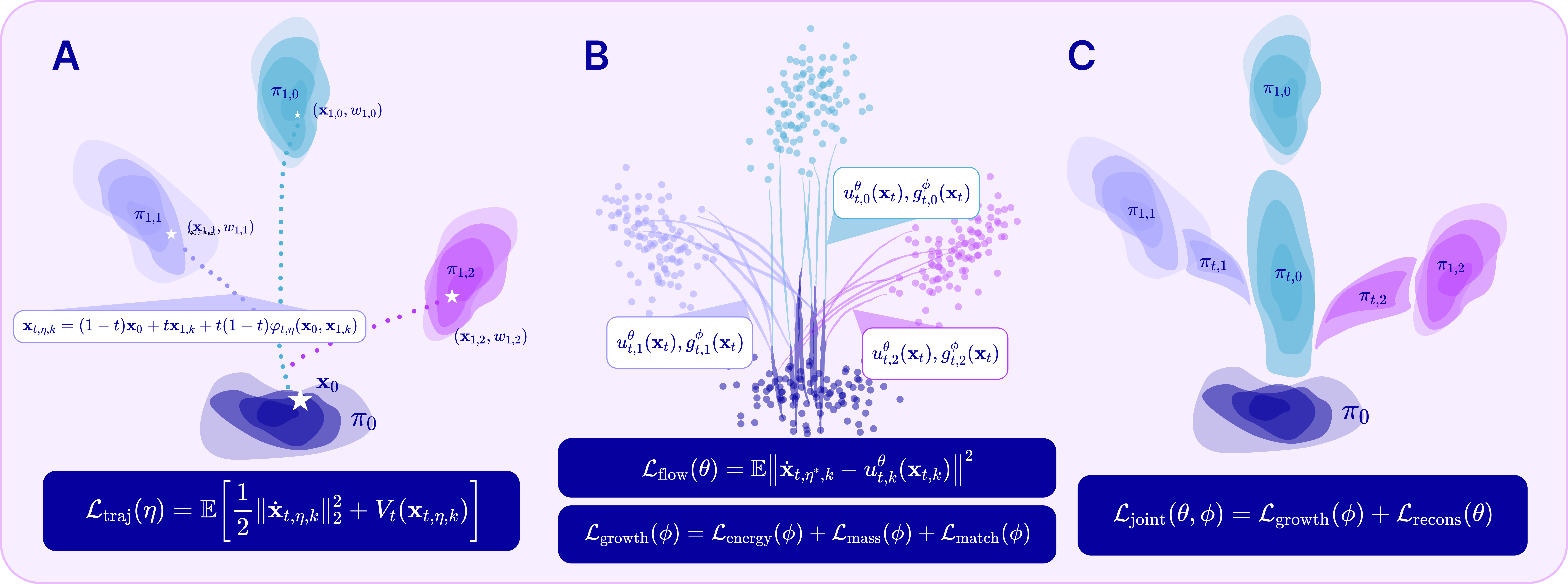
| Nov 11, 2025 | EntangledSBM is out! Read our paper on simulating interacting multi-particle systems with a novel Schrödinger bridge matching framework! |
|---|---|
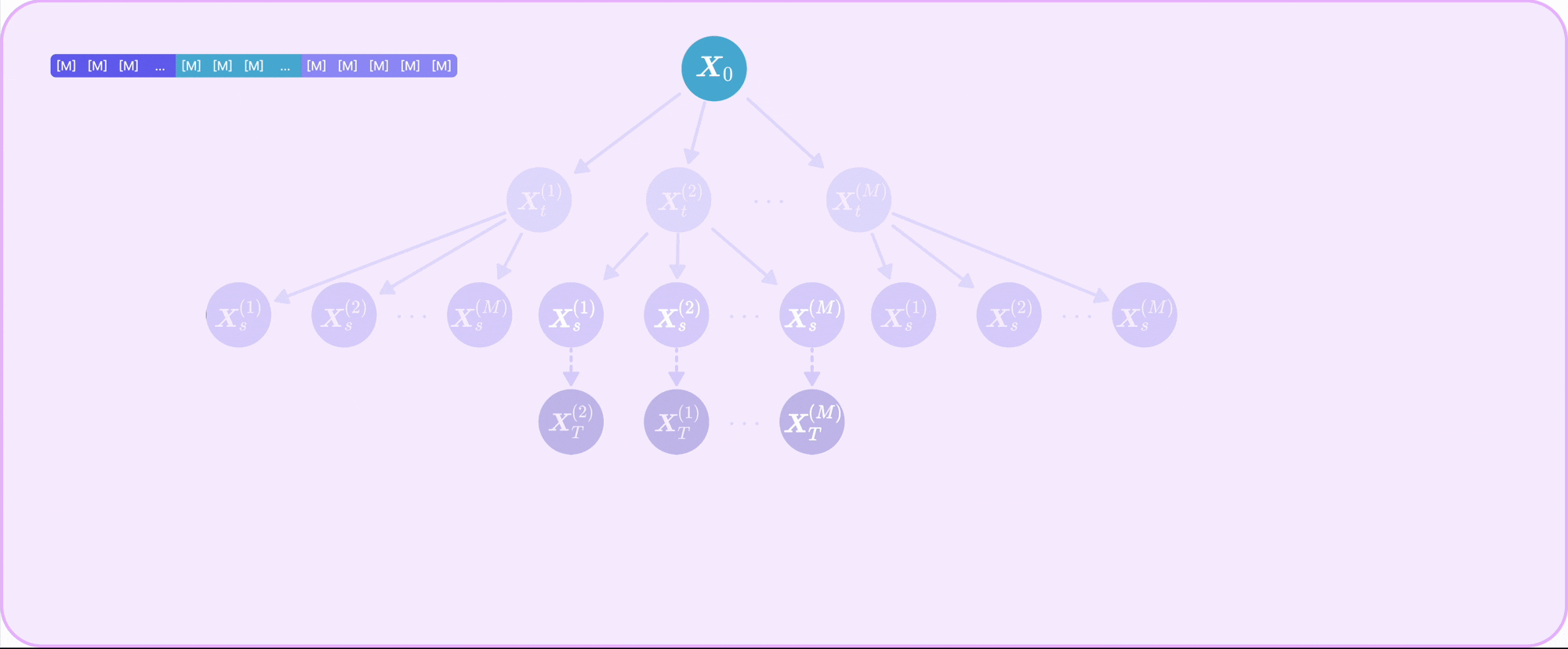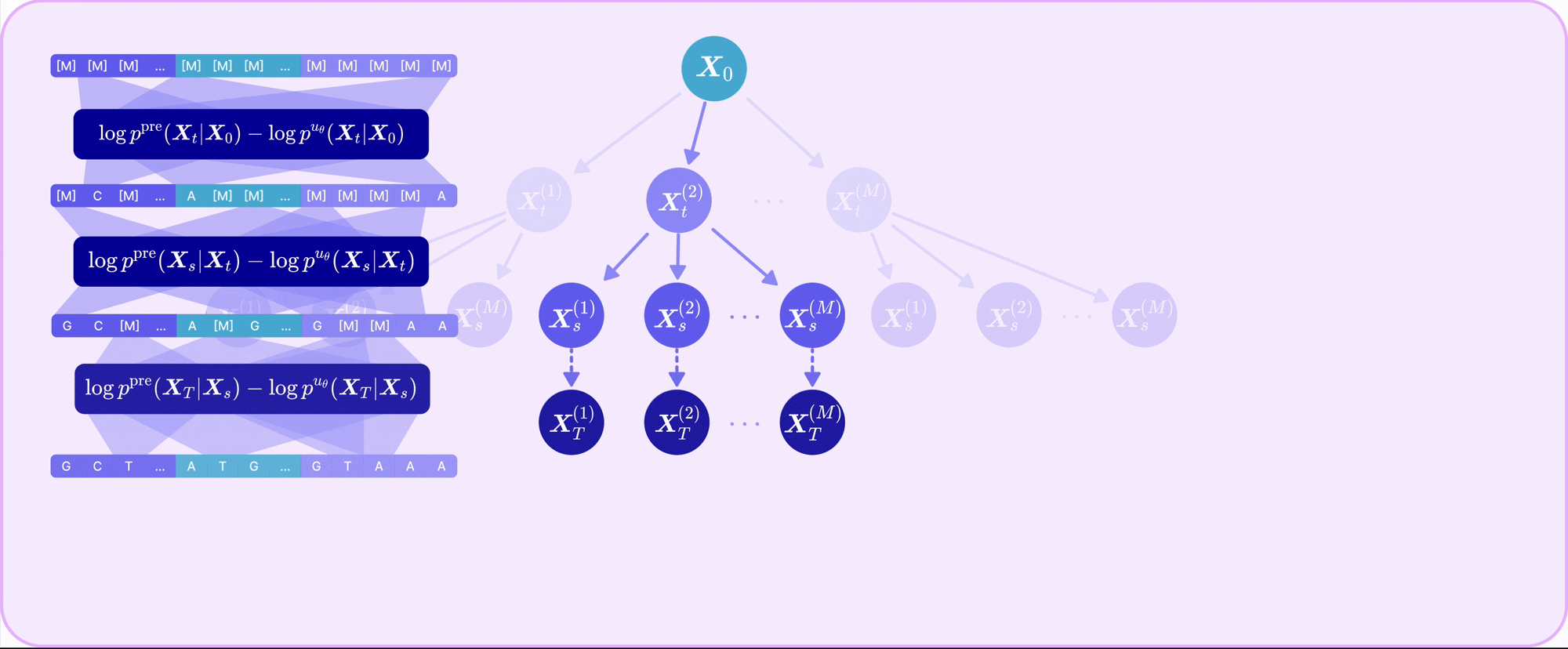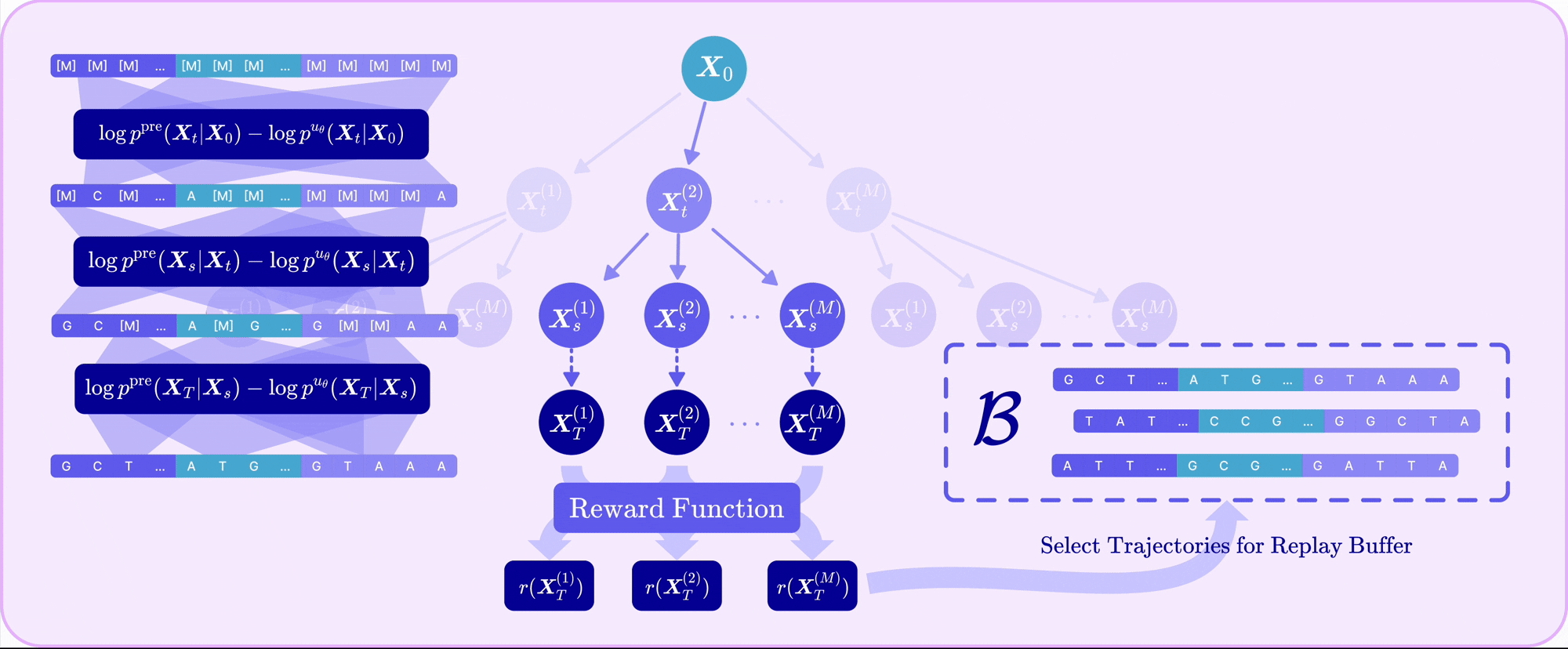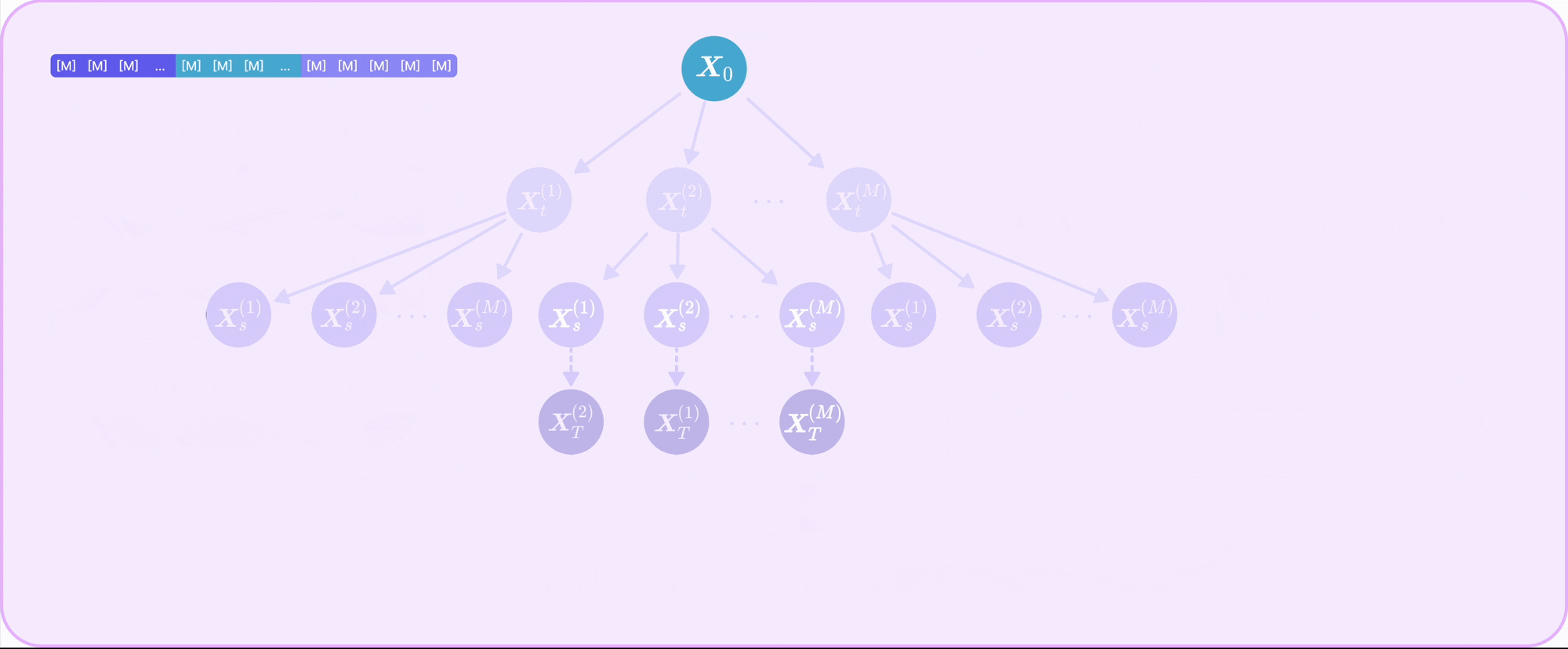
| Sep 30, 2025 | TR2-D2 is out! Read our paper on off-policy RL for discrete diffusion fine-tuning with multi-objective rewards! |
| Jun 11, 2025 | Branched Schrödinger Bridge Matching released on arXiv and accepted at ICML ExAIT workshop! Come by our poster in Vancouver! |
| May 01, 2025 | PepTune: De Novo Generation of Therapeutic Peptides with Multi-Objective-Guided Discrete Diffusion accepted at ICML 2025! Come by us at Poster Session 4 to chat (+ free PepTune stickers) |
Selected Publications
For a full list, see my publications page or google scholar profile.